Razvan Marinescu
Assistant Professor
UC Santa Cruz
ramarine at ucsc.edu
Prospective PhDs/postdocs/undergrads/masters who want to work with me: see here
I am an Assistant Professor in the department of Computer Science and Engineering at UC Santa Cruz. My research focuses on Machine Learning and its applications to Healthcare and Biomolecular systems. I am also the co-founder and CTO of a drug screening start-up, GiwoTech Inc.
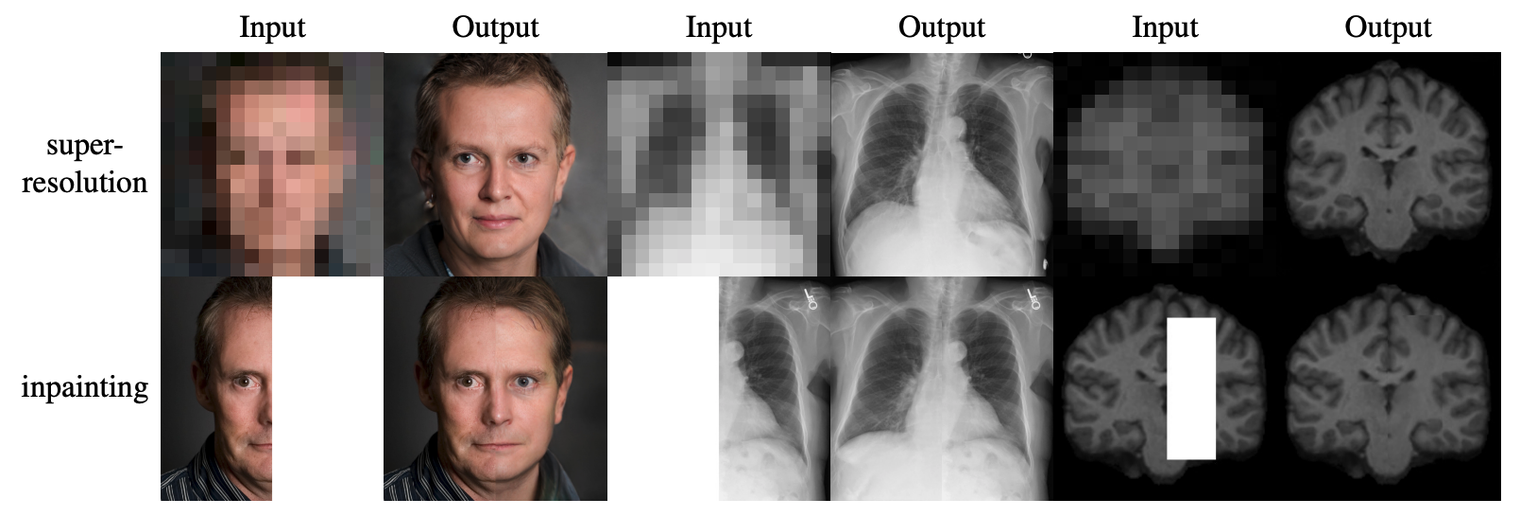
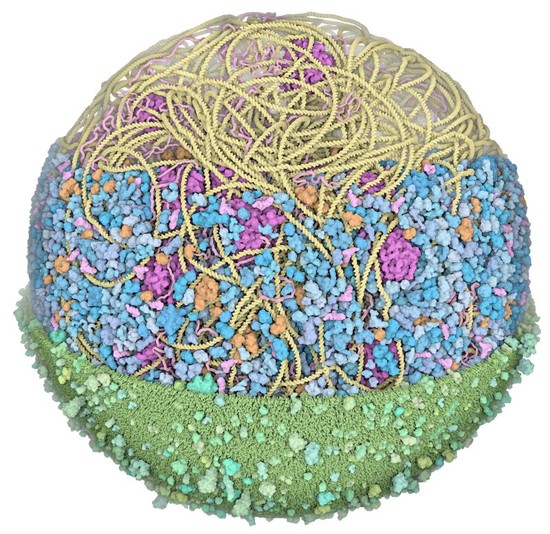
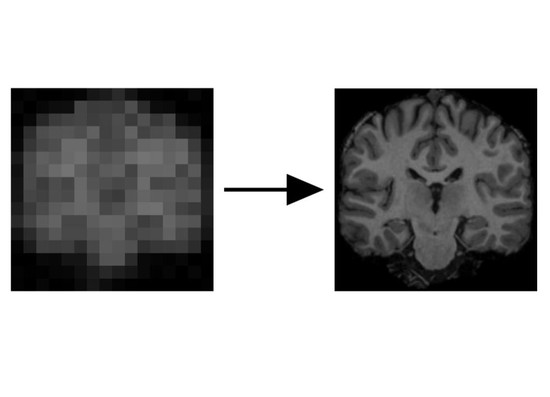
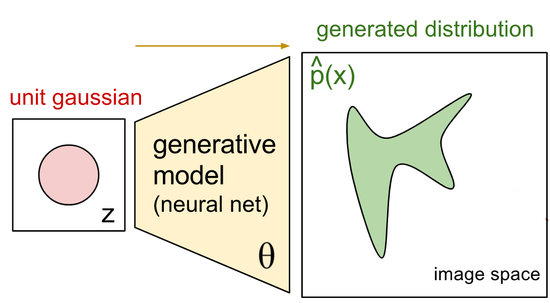
During my PostDoc in Polina Golland’s lab at MIT, I worked on machine learning algorithms for healthcare applications. I developed generative models for natural images, Chest X-Rays and brain images, which can be used as prior models for image reconstruction tasks, using bayesian posterior optimisation.
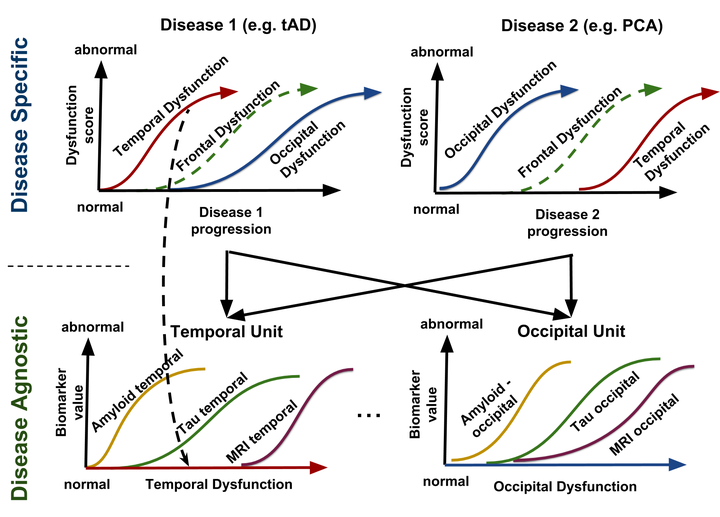
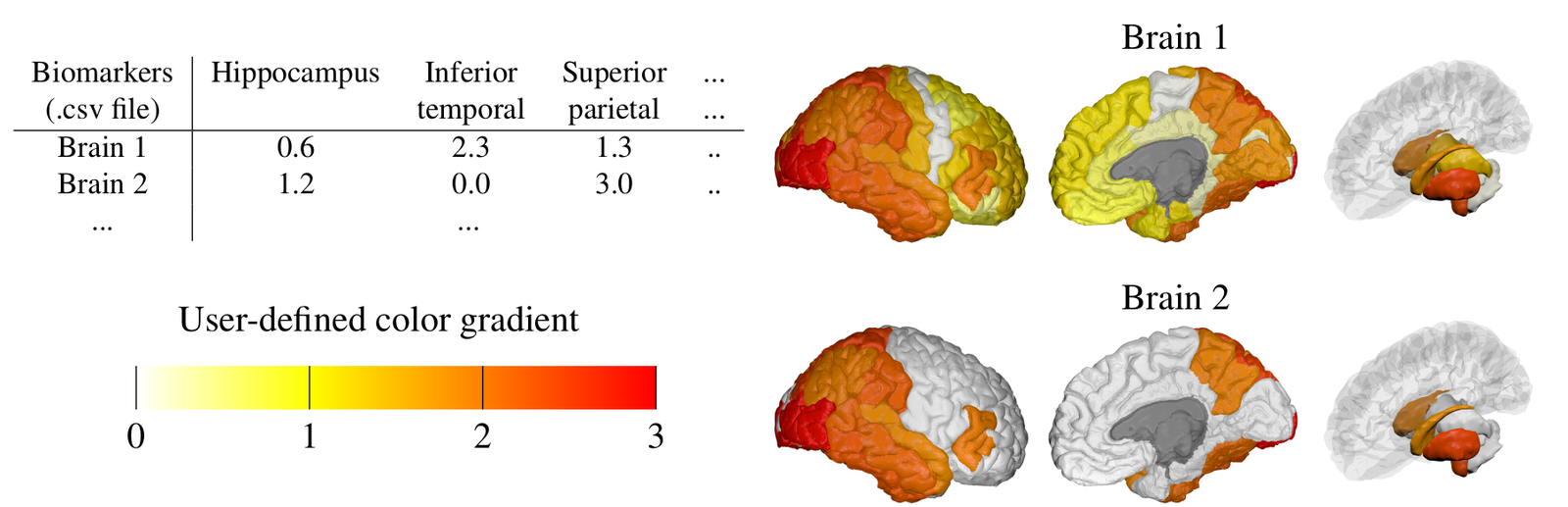
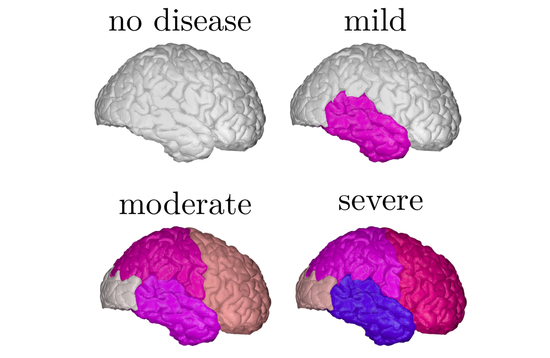
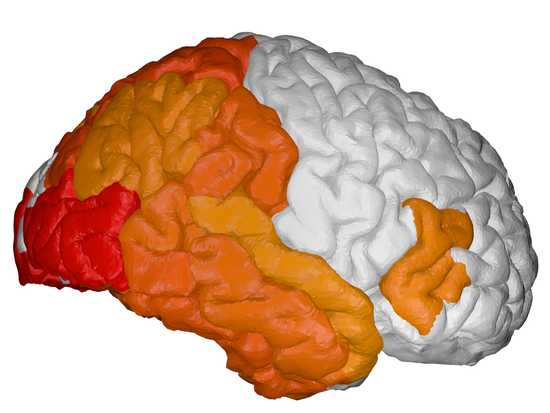
During my PhD, I developed Bayesian statistical models for prediction of Alzheimer’s disease and related neurodegenerative diseases. Such models were able to not only to estimate the continuous evolution of Alzheimer’s disease, but also discover novel spatial patterns of brain pathology (e.g. DIVE) or transfer such “learned evolutions” in populations with different diseases (e.g. DKT). I further co-organised TADPOLE, an international competition that compared 92 algorithms from 33 teams at predicting the evolution of Alzheimer’s disease. Finally, together with UCL colleagues N. Firth and S. Primativo, I performed one of the first comprehensive clinical studies (Firth et al., Brain, 2019) on the evolution of Posterior Cortical Atrophy, a rare dementia affecting the visual system.
On the technical side, I particularly enjoy the mathematics of statistical inference methods for Machine Learning. For example, I derived the EM-update rules for two complex Bayesian models that I proposed (see Supplementary section of the DIVE article and Appendix sections B and D of my PhD thesis).
I also have a keen interest in general machine learning and computational intelligence, in order to make computers perform intelligent tasks. Aside from research, at MIT I was also the president of the Postdoctoral Association, working with the MIT administration to create a better research environment for postdocs.
You can see a complete list of publications on my Google Scholar page. See my academic job market research statement here.
My wife Leilani Gilpin is also a researcher in ML, and does fantastic work on ML Explainability using logical reasoning methods. See her publications here.
Interests
- Machine Learning
- Generative ML Models
- Bayesian inference
- Applications of Machine Learning to Medicine
- Molecular Biology
- Molecular Dynamics
- Entrepreneurship
Education
-
PostDoc in Polina Golland's lab
MIT CSAIL
-
PhD in Computer Science, 2019
Center for Medical Image Computing, UCL
-
MEng in Computer Science, 2014
Imperial College London
-
BSc in Computer Science, 2013
Imperial College London