Abstract
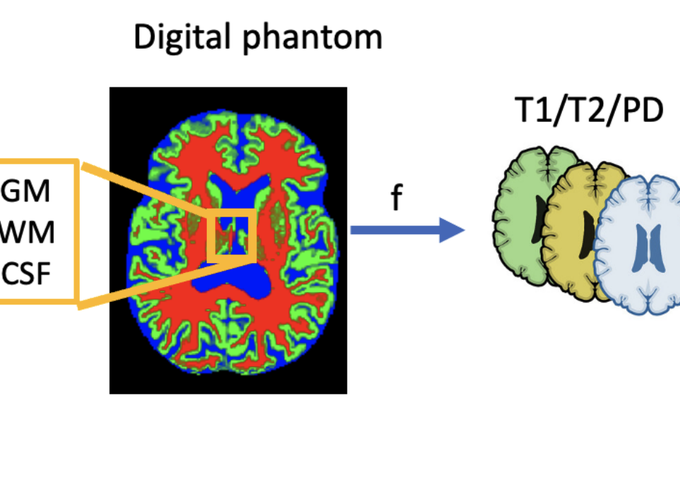
Reconstructing digital brain phantoms in the form of voxel-based, multi-channeled tissue probability maps for individual subjects is essential for capturing brain anatomical variability, understanding neurological diseases, as well as for testing image processing methods. We demonstrate the first framework that estimates brain tissue probability maps (Grey Matter - GM, White Matter - WM, and Cerebrospinal fluid - CSF) with the help of a Physics-based differentiable MRI simulator that models the magnetization signal at each voxel in the volume. Given an observed T1/T2-weighted MRI scan, the corresponding clinical MRI sequence, and the MRI differentiable simulator, we estimate the simulator’s input probability maps by back-propagating the L2 loss between the simulator’s output and the T1/T2-weighted scan. This approach has the significant advantage of not relying on any training data and instead uses the strong inductive bias of the MRI simulator. We tested the model on 20 scans from the BrainWeb database and demonstrated a highly accurate reconstruction of GM, WM, and CSF. Our source code is available online: https://github.com/BioMedAI-UCSC/BMapEst.